Cucumis melo

Mature melons. Image provided by J. McCreight, USDA-ARS, Salinas California.
- Melon Breeding and Genetics
- Fusarium wilt
- Powdery Mildew
- Cucumber mosaic virus (CMV)
- Cucurbit yellow stunting disorder virus (CYSDV)
Melon Breeding and Genetics
The center of origin of melon remains unsettled among the melon genetic and breeding community. Taxonomists regarded Asia as the center of origin and domestication for melon based on the rich phenotypic diversity of melons across the southern portion of the continent. Subsequent comparisons of chromosomes among various Cucumis species pointed to Africa as the center origin despite the paucity of diversity of melons from that continent. Recent analyses of molecular and other data of samples from across the natural geographic range of Cucumis (Africa, southern and eastern Asia and Australia) suggest India as the center of origin for melon.
Draft genome sequences for melon (Cucumis melo) were established in 2012. Breeding for specifics disease resistances in melon are underway in labs, greenhouses and fields in South Carolina, California and Arizona. A Melon Core collection of 384 lines will be established
DNA of available melon accessions in the U.S. National Plant Germplasm (2038 accessions including some heirlooms) has been sampled from greenhouse-grown plants. A Melon Core collection will be established based on the GBS analyses. Seed of the accessions included in the Melon Core will be produced for distribution through USDA-NPGS-GRIN.
Fusarium wilt
Fusarium wilt of cucurbits, caused by the fungal pathogen Fusarium oxysporum, is one of the most devastating of all cucurbit soil-borne diseases. Fusarium wilt of melon is caused by Fusarium oxysporum f. sp. melonis (FOM), and is considered one of the more important diseases of melon in the United States (Wechter et al., 1995; Zuniga et al., 1997). This pathogen also causes severe losses in a number of other countries, including those in South America (Erzurum et al., 1999) and Europe (Belisario et al., 2000), Republic of South Africa (Schreuder et al., 2000), and Japan (Namiki et al., 2000). There are currently no economical or even viable chemical control strategies, or other methods that can control this soil-borne pathogen. Four races of FOM have been described in melon: 0, 1, 2, and 1,2 based on resistance genes. Two variants of race 1,2 have been identified: 1,2y, which induces yellowing reaction, and 1,2w, which induces wilting response (Zink and Thomas, 1990). Genetic studies on inheritance of resistance to the different races of Fom (Risser et al., 1976; Zink and Thomas, 1990), as well as identification of genetic markers associated with these resistances have been described (Ficcadenti et al., 2002; Joobeur et al., 2004; Tezuka et al., 2009; Wang et al., 2000; Wechter et al., 1998; Wechter et al., 1995; Zheng et al., 1999). Joobeur et al., (2004) identified and described the Fom-2 locus, which confers resistance to Fom race 1. Tezuka et al., (2009) mapped the Fom-1 locus, which confers resistance to race 2. No reports have been published to date of QTL analysis for additional, minor gene loci involved in FOM resistance to these two races.
PI 124111 and its derived line, MR-1, are highly resistant to FOM races 0, 1, and 2. MR-1 also possesses significant resistance to a number of other diseases, including powdery mildew, Alternaria leaf blight and downy mildew (Thomas, 1982; Thomas, 1985; Thomas, 1986). The unique resistance qualities of MR-1 make it an extremely important line for studies of resistance and as a donor for introgression of resistances into susceptible lines and cultivars. MR-1 was crossed with the sweet Israeli cultivar ‘Ananas Yok’neam’ (AY). AY is susceptible to many plant pathogens including FOM, and the causal agents of downy mildew, powdery mildew, and Alternaria leaf blight(Thomas, 1982; Thomas, 1985; Thomas, 1986). The cross of MR-1 and AY initiated transfer of FOM resistance to sweet melons. A recombinant inbred line (RIL) population, derived by single seed descent from the F2 population was developed from this cross. Two hundred RILs F8 to F10 have been generated for molecular studies and breeding work associated with this project. A subset of this RIL population was used to identify QTL associated with Alternaria leaf blight resistance (Daley et al., 2017). This population is being evaluated for resistance to FOM races 1 and 2 in Charleston, SC.
Breeding objectives
- Introgress resistance to FOM races 1 and 2 into melon germplasm acceptable and adapted for production in the eastern and western U.S. melon production areas.
- Identify QTL for resistance to FOM races 1 and 2.
Powdery Mildew
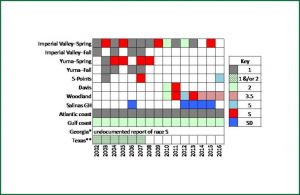
Powdery Mildew (PM; incited on melon in the U.S. by Podosphaera xanthii) is a constant threat to melon production in every growing region. Resistance breeding was initiated by USDA in the 1920s (Whitaker and Jagger, 1937) and continues through the present due to the emergence of new races that overcome resistance genes (McCreight et al., 2012). Melon powdery mildew races 1 and 2 are important in the eastern U.S. (C.S. Kousik, unpublished) and Texas (K.M. Crosby, personal communication). Races 1 and 2 occur in California and Arizona, but three other races are known there as well: 3.5 (Woodland-Davis, CA), 5 (Five Points, CA), and S (Imperial Valley, CA and Yuma, AZ) (McCreight, 2002). A survey of powdery mildew races in California was done in 2015 and 2016. Read more about Cucurbit Powdery mildew at the Melon Powdery Mildew Database at University of California Riverside.
PI 124111 exhibits high-level resistance to PM races 1, 2, 3, 4, 5, and twelve Czech races, but is susceptible to races 3.5, S, and SD (McCreight et al., 2012). Melon breeding line MR-1 was selected from PI 124111 for uniform reaction to downy and powdery mildews; when released it was known to be resistant to powdery mildew races 1, 2, and 3 (Thomas, 1986). A recombinant inbred line (RIL) population from a cross of MR-1 with the powdery mildew susceptible cultivar Ananas Yok’neam (AY) has been advanced to the F8 to F10 generation. This population is being evaluated for resistances to races 1 and 2 in Charleston, SC, and race 5 in Five Points, CA. Genetic analyses (Mendelian and QTL) will be done for resistances to these three races. Selected multiple race-resistant RILs will be introgressed to western U.S. shipper type and eastern type cantaloupes.
PI 313970 exhibits high-level resistance to powdery mildew races 1, 2, 3.5, 4.5, and S (McCreight, 2003) , as well as resistance or susceptibility to thirteen Czech races, and is susceptible to race SD (McCreight and Coffey, 2011; McCreight et al., 2012; Pitrat and Besombes, 2008). A RIL population from a cross of PI 313970 with ‘Top Mark’ is being developed and will be characterized primarily for resistances to races 1, 2, and S, but may also be characterized for reactions to races 3.5 (Woodland, CA) and 5 (Five Points, CA).
- First view of race S in a commercial melon field near Holtville, CA, Spring 2003 (McCreight et al., 2005). Image provided by Jim McCreight, USDA-ARS.
- U.S. melon powdery mildew races summarized from published and unpublished data. Image provided by J. McCreight, USDA-ARS.
- ‘Golden Beauty Casaba’ plants sustaining severe powdery mildew infection that resulted in loss of foliage, which reduces lower sugar content of the fruit; loss of foliage also exposes the fruit to direct sunlight, which leads to fruit quality loss from sunburn. Image provided by Jim McCreight, USDA-ARS.
- Heavy powdery mildew infection on melons, where abaxial and adaxial surfaces are covered with mycelia spores. Image provided by Jim McCreight, USDA-ARS.
- Under “ideal” powdery mildew conditions, crowns and internodes will be infected. Image provided by Jim McCreight, USDA-ARS.
- Race 1 test of MR-1 x AY recombinant inbred lines (RIL) in a greenhouse, Charleston, SC. Image provided by Shaker Kousik, USDA-ARS.
Cucumber mosaic virus (CMV)
Cucumber mosaic virus (CMV) is distributed world-wide and can occur wherever cucurbits are cultivated. The virus is transmitted by more than 80 species of aphid (Gallitelli , 2000), and can be acquired and transmitted during brief, superficial probes of plants (Martin et al., 1997). CMV has an extensive and broad host range among crops and weeds, including more than 1200 species in 100 plant families (Zitter and Murphy, 2009). CMV has affected melon in the U.S. for more than 100 years, including Yuma, AZ and the Central Valley of Arizona (Coudriet, 1962; Gilbert, 1916). Cucurbit hosts differ in their susceptibility to CMV, with melon, cucumber, pumpkin and squash affected more severely by the virus than watermelon. Foliar symptoms on infected cucurbit plants include yellow spotting (Fig. 1), downward bending of petioles, and curling of leaves. As symptom development progresses, plants develop mosaic symptoms (Fig. 2), more extensive leaf curling, and sometimes leaf deformation. Fruit symptoms vary among cucurbits as well as by genotype, ranging from asymptomatic to severe rugosity (roughness), and occasional color breaking. Fruit quality and marketability are often reduced (Zitter and Murphy, 2009). Symptoms on cucurbit plants can resemble those caused by viruses of other genera, particularly those of the genus Potyvirus; therefore confirmation with serological or molecular methods is needed for correct identification.
The genome of CMV consists of three separate RNA molecules approximately 29 nm in diameter, each packaged separately into virus particles (virions) (Lot and Kaper, 1976). CMV symptom severity can be influenced by satellite RNA molecules that become associated with some CMV isolates. These satellite RNAs are fully dependent on CMV for replication, and they are packaged in CMV virions and can be transmitted plant-to-plant by aphid vectors along with the virus. The presence of satellite RNAs with CMV can lead to increased or decreased symptom severity (Francki 1985; Palukaitis, 1988).

Initial symptoms of leaf spotting caused by CMV infection of watermelon. Image provided by Bill Wintermantel, USDA-ARS.
Resistance in ‘Freeman cucumber’ (C. melo Group Conomon) was reported to be controlled by three recessive genes that resulted in lower virus titer than in the susceptible parent, the honeydew ‘Noyamid’ (C. melo Group Inodorus); the F1 was intermediate for virus titer (Karchi et al., 1975). Breeding for resistance to CMV is challenged by numerous strains able to overcome resistance genes that provide resistance to other strains (Lin et al., 2003). There are as of yet no commercially available CMV-resistant (CMVR) melon cultivars adapted to the western production areas. The California Melon Research Board supported a breeding project at Cornell in the 1990s to develop western U.S. shipper type melons (cantaloupe and honeydew) that culminated in the release of CMV-tolerant (CMVT) breeding lines selected for lack of symptoms and/or marketable fruit production despite foliar symptoms.
Pedigrees of the CMVT Cornell melons are diverse, with tolerance derived from combinations of the following three sources: Freeman cucumber and PI 161375 (Group Chinensis), which are tolerant to CMV and Watermelon mosaic virus (Enzie, 1943; Pitrat, 1978), and KCMR, which was received by Dr. H.M. Munger in 1978 from Z. Karchi (ARO, Neve Ya’ar, Israel).

Mosaic symptoms caused by CMV infection of melon. Image provided by Bill Wintermantel, USDA-ARS.
Breeding objectives
- Increase Cornell CMVT lines for assessment of fruit quality and level of tolerance, as indicated by virus titer. Virus reaction tests will include two or more strains of CMV.
- Evaluate performance of Cornell CMVT lines in controlled-inoculation greenhouse tests and open field trials in areas where CMV is likely to occur, based in input from CucCAP stakeholders. They will also be assessed for adaptation and fruit quality in field tests at three locations in Arizona and California. We will also test the putative commercially developed CMVR lines in these tests.
- Identify QTL for CMV resistance.
- Identify molecular markers for CMV resistance.
Cucurbit yellow stunting disorder virus (CYSDV)
Melon production is threatened by the crinivirus CYSDV in areas favorable to its vector, the sweetpotato whitefly, Bemisia tabaci Gennadius. At least three cryptic species of B. tabaci: NW1, MEAM1, and MED (formerly known as biotypes A, B, and Q, respectively) have been shown to transmit the virus (Célix et al., 1996; Wisler et al., 1998), although it is likely other cryptic species of B. tabaci can transmit CYSDV as well. The virus is not transmitted mechanically, by seed, or by other known methods.
The most recognizable symptoms of CYSDV include leaf yellowing and chlorosis between the veins (Fig. 1), but early symptoms can also include chlorotic mottle (Fig. 2). Symptoms first appear on older leaves about three weeks after initial infection, and spread acropetally (Fig. 3) (Célix et al., 1996; Wintermantel et al., 2009). Infected plants may become completely chlorotic (Fig. 4). There is a latent period of about three weeks during which CYSDV-infected plants are asymptomatic, but the virus is increasing in titer and moving through the vascular system from which it can be acquired and serially transmitted by whiteflies to new growth of infected plants as well as to neighboring plants It is, therefore, not recommended to move cucurbits from areas with active CYSDV infections even if plants are not exhibiting symptoms.
- Figure 3. CYSDV-infected plant exhibiting typical pattern of symptom expression that appears at the crown and develops acropetally, and in the case of early infection can result in 100 % yellowing of the of the plant. The older leaves are often thinned and brittle. Image provided by Jim McCreight, USDA-ARS.
- Figure 4. Extreme, nearly 100% yellowing due to CYSDV infection. Image provided by Jim McCreight, USDA-ARS.
- Figure 5. Naturally infected field tests for CYSDV resistance in melon. Left: overview of PI 313970 and selections from crosses of PI 313970 with susceptible melons. Right: melon accession screening tests for new sources of resistance to the virus. Image provided by Jim McCreight, USDA-ARS.
CYSDV first appeared on melon in 1982 in the United Arab Emirates (Hassan and Duffus, 1991) and then spread throughout the Mediterranean basin, reaching Spain by 1989 (Célix et al., 1996; Sese et al., 1994). The virus appeared in the lower Rio Grande area of Texas and the adjoining area of Mexico in 1999 (Kao et al., 2000) and later emerged in lower deserts of the southwestern U.S. (Yuma and central Arizona, and Coachella, Imperial and Palo Verde Valleys of California) and western Mexico during the Fall season of 2006 (Wintermantel et al., 2009) followed by Florida in 2007 (Polston et al., 2008). The virus has also been reported in China (Liu et al., 2010).
CYSDV can infect and cause disease symptoms on many members of the Cucurbitaceae, including melon, watermelon, squash, pumpkin and some wild cucurbits (Wintermantel et al., 2009). The virus also infects a number of weeds common to regions where melons are produced, and these non-cucurbit host plants vary as sources for transmission of CYSDV to cucurbit hosts by whiteflies (Wintermantel et al., 2016). Most geographically distant isolates of CYSDV are highly conserved genetically, suggesting recent spread of the virus in most regions, however a divergent Eastern group of isolates from Saudi Arabia, Iran, and Sudan has been identified as well (Mohammed et al., 2014; Rubio et al., 2001; Yakoubi et al., 2007).
Resistance to CYSDV in melon was first identified in Spain in TGR 1551 (PI 412420) and TGR 1937 (PI 482431) (López-Sesé and Gómez-Guillamón, 2000). Resistance was observed in PI 313970 when the virus first appeared in California in 2006 (Fig. 5) (McCreight and Wintermantel, 2011). Several other sources of resistance to CYSDV have been identified, and all appear to be conditioned by recessive genes (McCreight et al., 2015; McCreight et al., 2016; McCreight et al., 2017)
Breeding objectives
- Develop CYSDV-resistant U.S. western shipping type cantaloupe and honeydew breeding lines.
- Identify QTL for CYSDV resistance in PI 313970.
Literature Cited
- Belisario, A., L. Luongo, L. Corazza, and T.R. Gordon. 2000. Indagini su popolaziono di Fusarium oxysporum f. sp. melonis in Italia. Colture Protette 3:87–80.
- Célix, A., A. López-Sesé, N. Almarza, M.L. Gómez-Guillamón, and E. Rodríguez-Cerezo. 1996. Characterization of Cucurbit yellow stunting disorder virus, a Bemisia tabaci-transmitted closterovirus. Phytopathology 86:1370-1376.
- Coudriet, D.L. 1962. Efficiency of various insects as vectors of Cucumber mosaic and Watermelon mosaic viruses in cantaloupes. J. Econ. Entomol. 55:519–520.
- Daley, J., S. Branham, A. Levi, R. Hassell, and P. Wechter. 2017. Mapping resistance to Alternaria cucumerina in Cucumis melo. Phytopathology. http://dx.doi.org/10.1094/PHYTO-06-16-0246-R
- Enzie, W.D. 1943. A source of muskmelon mosaic resistance found in the oriental pickling melon, Cucumis melo var. conomon. Proc. Amer. Soc. Hort. Sci. 43:195–198.
- Erzurum, K., Y. Taner, E. Secur, R. Yanmaz, and S. Maden. 1999. Occurrence of races of Fusarium oxysporum f. sp. melonis causing wilt on melon in Central America.
- Ficcadenti, N., S. Sestili, S. Annibali, G. Campanelli, A. Belisario, M. Maccaroni, and L. Corazza. 2002. Resistance to Fusarium oxysporum f. sp. melonis race 1,2 in muskmelon lines Nad-1 and Nad-2. Plant Dis. 86:897–900.
- Francki, R.I.B. 1985. Plant virus satellites. Ann. Rev. Microbiol. 39: 151–174.
- Gallitelli, D. 2000. The ecology of Cucumber mosaic virus and sustainable agriculture. Virus Res. 71: 9–21.
- Gilbert, W.W. 1916. Cucumber mosaic disease. Phytopathology 6:143–145.
- Hassan, A.A. and J.E. Duffus. 1991. A review of a yellowing and stunting disorder of cucurbits in the United Arab Emirates. Emirates J. Agr. Sci. 2:1-16.
- Joobeur, T., J.J. King, S.J. Nolin, C.E. Thomas, and R.A. Dean. 2004. The Fusarium wilt resistance locus Fom-2 of melon contains a single resistance gene with complex features. Plant J. 39:283–297.
- Kao, J., L. Jia, T. Tian, L. Rubio, and B.W. Falk. 2000. First report of Cucurbit yellow stunting disorder virus (genus Crinivirus) in North America. Plant Dis. 84:101.
- Karchi, Z., S. Cohen, and A. Govers. 1975. Inheritance of resistance to Cucumber mosaic virus in melons. Phytopathology 65:479–481.
- Lin, H.X., L. Rubio, A. Smythe, M. Jiminez, and B.W. Falk. 2003. Genetic diversity and biological variation among California isolates of Cucumber mosaic virus. J. Gen. Virol. 84:249–258.
- Liu, L.Z., Y.Y. Chen, and W.M. Zhu. 2010. First report of Cucurbit yellow stunting disorder virus on melon in China. Plant Dis. 94:485.
- López-Sesé, A.I. and M.L. Gómez-Guillamón. 2000. Resistance to Cucurbit yellowing stunting disorder virus (CYSDV) in Cucumis melo L. HortScience 35:110–113.
- Lot, H. and Kaper, J.M. 1976. Further studies on the RNA component distribution among the nucleoproteins of Cucumber mosaic virus. Virology 74: 223–226.
- Martin, B., Collar, J.L., Tjallingii, W.F., and Fereres, A. Intracellular ingestion and salivation by aphids may cause the acquisition and inoculation of non-persistently transmitted plant viruses. J. Gen. Virol. 78: 2701–2705.
- McCreight, J.D. 2002. Powdery mildew race 1 in Imperial Valley, California. Cucurbit Genet. Coop. Rpt. 25:20–21.
- McCreight, J.D. 2003. Genes for resistance to powdery mildew races 1 and 2U.S. in melon PI 313970. HortScience 38:591–594.
- McCreight, J.D. and M.D. Coffey. 2011. Inheritance of resistance in melon PI 313970 to cucurbit powdery mildew incited by Podosphaera xanthii race S. HortScience 46:838–840.
- McCreight, J.D., M.D. Coffey, T.A. Turini, and M.E. Matheron. 2005. Field evidence for a new race of powdery mildew on melon. HortScience 40(4):888 (abstr.).
- McCreight, J.D., M.D. Coffey, B. Sedlakova, and A. Lebeda. 2012. Cucurbit powdery mildew of melon incited by Podosphaera xanthii: Global and western U.S. perspectives, p. 181–189. In: N. Sari, I. Solmaz, and V. Aras (eds.). Cucurbitaceae 2012, Proceedings of the Xth EUCARPIA meeting on genetics and breeding of Cucurbitaceae, Antalya (Turkey), October 15-18th, 2012.
- McCreight, J.D. and W.M. Wintermantel. 2011. Genetic resistance in melon PI 313970 to Cucurbit yellow stunting disorder virus. HortScience 46:1582–1587.
- McCreight, J.D., W.M. Wintermantel, and E.T. Natwick. 2015. Inheritance of resistance to Cucurbit yellow stunting disorder virus (CYSDV) in melon (Cucumis melo L.) accessions PI 482431 (TGR 1937) and PI 614479. Proceedings IXVIII. International Plant Protection Congress, 24–27 August 2015, Berlin.
- McCreight, J.D., W.M. Wintermantel, and E.T. Natwick. 2016. New Sources of Resistance to CYSDV in Melon, p. 61–65. In: E.U. Kozik, and H.S. Paris (eds.). Cucurbitaceae 2016, XIth Eucarpia Meeting on Genetics and Breeding of Cucurbitaceae, Warsaw, Poland.
- McCreight, J.D., W.M. Wintermantel, E.T. Natwick, J.W. Sinclair, K.M. Crosby, and M.L. Gómez-Guillamón. 2017. Recessive resistance to Cucurbit yellow stunting disorder virus in melon TGR 1551. Acta Hort. Accepted 24 Nov. 2015 (In Press).
- Mohammed, H.S., S. Zicca, A. Manglli, M.E. Mohamed, M.A. El Siddig, L. Tomassoli, and A.A. El Hussein. 2014. Identification and phylogenetic analysis of common pumpkin viruses in Sudan. . J. Plant Pathol. 96:77–84.
- Namiki, F., K. Shimizu, K. Satoh, T. Hirabayashi, K. Nishi, T. Kayamura, and T. Tsuge. 2000. Occurrence of Fusarium oxysporum f. sp. melonis race 1 in Japan. J. Gen. Plant Pathol. 66:12–17.
- Polston, J.E., L.L. Hladky, F. Akad, and W.M. Wintermantel. 2008. First report of Cucurbit yellow stunting disorder virus in cucurbits in Florida. Plant Dis. 92:1251.
- Palukaitis, P. 1988. Pathogenicity regulation by satellite RNAs of cucumber mosaic virus: minor nucleotide sequence changes alter host responses. Mol. Plant-Microbe Interactions. 1: 175–181.
- Pitrat, M. and D. Besombes. 2008. Inheritance of Podosphaera xanthii resistance in melon line ‘90625’, p. 135–142. In: M. Pitrat (ed.). Cucurbitaceae 2008, IXth EUCARPIA Meeting on Genetics and Breeding of Cucurbitaceae. INRA, Avignon, France.
- Pitrat, M. 1978. Tolerance of melon to Watermelon mosaic virus II. Cucurbit Genet. Coop. Rpt. 1:20.
- Risser, G., Z. Banihashemi, and D.W. Davis. 1976. A proposed nomenclature of Fusarium oxysporum f.sp. melonis races and resistance genes in Cucumis melo. Phytopathology 66:1105–1106.
- Rubio, L., Y. Abou-Jawdah, H.-X. Lin, and B.W. Falk. 2001. Geographically distant isolates of the crinivirus Cucurbit yellow stunting disorder virus show very low genetic diversity in the coat protein gene. J. Gen. Virol. 82:929–933.
- Schreuder, W., S.C. Lamprecht, and G. Holz. 2000. Race determination and vegetative compatibility grouping of Fusarium oxysporum f. sp melonis from South Africa. Plant Dis. 84:231–234.
- Sese, A.I.L., M.L. Gomez-Guillamon, and J.R. Diaz-Ruiz. 1994. Appearance of a possible new melon yellowing disease in Spain. Cucurbit Genet. Coop. Rpt. 17:72–73.
- Tezuka, T., K. Waki, K. Yashiro, M. Kuzuya, T. Ishikawa, Y. Takatsu, and M. Miyagi. 2009. Construction of a linkage map and identification of DNA markers linked to Fom-1, a gene conferring resistance to Fusarium oxysporum f.sp. melonis race 2 in melon. Euphytica 168:177–188.
- Thomas, C.E. 1986. Downy and powdery mildew resistant muskmelon breeding line MR-1. HortScience 21:329.
- Thomas, C.E. 1985. Resistant reaction of muskmelon line MR-1 against downy mildew. Phytopathology 75:504 (Abstr.).
- Thomas, C.E. 1982. Resistance to downy mildew in Cucumis melo plant introductions and American cultivars. Plant Dis. 66:500–502.
- Wang, Y.H., C.E. Thomas, and R.A. Dean. 2000. Genetic mapping of a Fusarium wilt resistance gene (Fom-2) in melon (Cucumis melo L.). Mol. Breeding 6:379–389.
- Wechter, W.P., R.A. Dean, and C.E. Thomas. 1998. Development of sequence-specific primers that amplify a 1.5-kb DNA marker for race 1 Fusarium wilt resistance in Cucumis melo L. HortScience 33:291–291.
- Wechter, W.P., M.P. Whitehead, C.E. Thomas, and R.A. Dean. 1995. Identification of a randomly amplified polymorphic DNA marker linked to the Fom 2 Fusarium wilt resistance gene in muskmelon MR-1. Phytopathology 85:1245–1249.
- Whitaker, T.W. and C.I. Jagger. 1937. Breeding and improvement of cucurbits, p. 207-232. Yearbook of Agriculture, U.S. Dept. Agric. U.S. Government Printing Office, Washington, D.C.
- Wintermantel, W.M., L.L. Hladky, A.A. Cortez, and E.T. Natwick. 2009. A new expanded host range of Cucurbit yellow stunting disorder virus includes three agricultural crops. Plant Dis. 93:685–690.
- Wintermantel, W.M., R.L. Gilbertson, J.D. McCreight, and E.T. Natwick. 2016. Host-specific relationship between virus titer and whitefly transmission of Cucurbit yellow stunting disorder virus. Plant Dis. http://dx.doi.org/10.1094/PDIS-11-14-1119-RE
- Wisler, G.C., J.E. Duffus, H.Y. Liu, and R.H. Li. 1998. Ecology and epidemiology of whitefly transmitted closteroviruses. Plant Dis. 82:270–280.
- Yakoubi, S., C. Desbiez, H. Fakhfakh, C. Wipf-Scheibel, M. Marrakchi, and H. Lecoq. 2007. Occurrence of Cucurbit yellow stunting disorder virus and Cucumber vein yellowing virus in Tunisia. J. Plant Pathol. 89:417-420.
- Zheng, X.Y., D.W. Wolff, S. Baudracco-Arnas, and M. Pitrat. 1999. Development and utility of cleaved amplified polymorphic sequences (CAPS) and restriction fragment length polymorphisms (RFLPs) linked to the Fom-2 Fusarium wilt resistance gene in melon (Cucumis melo L.). Theoretical Appl. Genet. 99:453–463.
- Zink, F.W. and C.E. Thomas. 1990. Genetics of resistance to Fusarium oxysporum f.sp. melonis races 0, 1, and 2 in muskmelon line MR-1. Phytopathology 80:1230–1232.
- Zitter, T. A., and Murphy, J. F. 2009. Cucumber mosaic virus. The Plant Health Instructor. DOI: 10. 1094/PHI-I-2009-0518-01.
- Zuniga, T.L., T.A. Zitter, T.R. Gordon, D.T. Schroeder, and D. Okamoto. 1997. Characterization of pathogenic races of Fusarium oxysporum f.sp. melonis causing Fusarium wilt of melon in New York. Plant Dis. 81:592–596.