Team members: Zhangjun Fei (Boyce Thompson Institute); Umesh Reddy (West Virginia St. Univ.); Amnon Levi (USDA, ARS); Yiqun Weng (USDA, ARS); Michael Mazourek (Cornell University); Pat Wechter (USDA, ARS); and Rebecca Grumet (Michigan State University) reported on their lab’s progress and plans.
Objectives Year 1: Develop common genomic approaches and tools for cucurbits.
Work in progress and plans
1.1. Develop genomic and bioinformatic platforms for cucurbit crops
1.1.1. Genotyping by sequencing
In closely working with Cornell Genomic Diversity Facility, we have set up the genotyping-by-sequencing (GBS) platform for the four cucurbit species: watermelon, melon, cucumber and squash. Due to the recent patent issue of the GBS technology, Cornell Genomic Diversity Facility has stopped GBS service for samples that are currently not registered in their system. Fortunately, this will not affect our proposed GBS of 10,000 cucurbit samples.
Meantime, the Mazourek lab is currently testing an alternative genotyping technology for cucurbit species. This genotyping strategy is based on sequencing variable regions of moderately repetitive elements (a few thousand copies per genome) and much cheaper and easier to work than GBS, but generates much less markers. It has been shown to work well in maize which has a highly repetitive genome. It will provide a nice alternative if it works in cucurbits.
1.1.2. Sequence data processing/analysis
We have evaluated and compared the performance of TASSEL-GBS (http://www.maizegenetics.net/#!tassel/c17q9) and GATK (https://www.broadinstitute.org/gatk/) in SNP calling using GBS data we have generated for C. maxima and C. moschata. SNPs called from the deep genome resequencing data (>60x genome coverage) of two individual plants (parents of the mapping populations) were used to serve as positive controls. Our results showed that GATK achieved higher accuracy in GBS SNP calling than TASSEL-GBS. More extensive and thorough evaluation of these two programs is underway. The obtained results will guide us to use more appropriate software and parameters in our future GBS data analysis.
1.1.3. ICuGI database development
We are in the process of re-implementing the ICuGI database using the GMOD Tripal system (http://gmod.org/wiki/Tripal) and the Chado database schema (http://gmod.org/wiki/Chado). Melon genome sequence has been processed and will be included in the database. Genome syntenies between watermelon, melon and cucumber have been identified and a genome syntenty browser will be implemented in the database using GBrowse_syn (http://gmod.org/wiki/GBrowse_syn). The newly implemented database is expected to go public this fall.
1.2. Perform GBS analysis of PI collections, establish core populations of the four species, and provide community resource for genome wide association studies (GWAS)
1.2.1. GBS of cucurbit species, establish molecular-informed core populations
(i) Establishment of high-throughput DNA isolation capacity
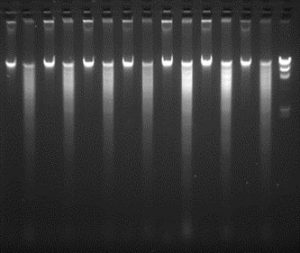 We have purchased equipment funded by Michigan State Univ. to establish a multiuser facility to facilitate high-throughput DNA isolation from samples in a 96 well format. This facility will serve as a resource for the full CucCAP project to prepare DNA for GBS on PI collections for the four cucurbit species. Protocols have been optimized to ensure high quality DNA preparation from cucurbit leaf samples, including both sample collection and preparation steps, and DNA isolation. Each crop team wishing to use the facility will grow the PI seedlings, sample leaf tissue in accordance with the optimized protocols and send the samples to the Grumet lab for processing. Given the extensive numbers of samples to be handled, it is important to ensure optimization, uniformity, and proper sample identity throughout the process. Quality of the DNA is verified according to the standards required by the Cornell Genomics Diversity Facility, including quantification, and gel analysis of uncut and HindIII digested DNA (300-500 ng/sample) as shown here for several cucumber samples.
We have purchased equipment funded by Michigan State Univ. to establish a multiuser facility to facilitate high-throughput DNA isolation from samples in a 96 well format. This facility will serve as a resource for the full CucCAP project to prepare DNA for GBS on PI collections for the four cucurbit species. Protocols have been optimized to ensure high quality DNA preparation from cucurbit leaf samples, including both sample collection and preparation steps, and DNA isolation. Each crop team wishing to use the facility will grow the PI seedlings, sample leaf tissue in accordance with the optimized protocols and send the samples to the Grumet lab for processing. Given the extensive numbers of samples to be handled, it is important to ensure optimization, uniformity, and proper sample identity throughout the process. Quality of the DNA is verified according to the standards required by the Cornell Genomics Diversity Facility, including quantification, and gel analysis of uncut and HindIII digested DNA (300-500 ng/sample) as shown here for several cucumber samples.
(ii) Sample collection, preparation and genotyping
The four crop team leaders have been working (or will work) with the germplasm curators to assemble a panel of diverse genotypes that includes materials in the PI collection at the National Plant Germplasm System (NPGS) as well as important open-pollinated or pureline cultivars. DNA will be sampled from a total of 1000-1600 accessions of each crop.
We have initiated DNA preparation from the cucumber PI collection. The first two plates have been sent to Cornell for GBS and sequencing data will be expected in the coming month. Data will be analyzed to assess the efficiency of the established high-throughput tissue sampling and DNA isolation protocol in GBS analysis.
